This tutorial is part of a series of R tutorials analyzing covid-19 data.
For part 1, see the following post:
https://www.sharpsightlabs.com/blog/r-data-analysis-covid-19-part1-data-wrangling/
Covid19 analysis, part 2: merge datasets
In this tutorial, we’re going to set up a process that will enable us to retrieve and wrangle some necessary covid19 datasets … data that we’ll need for further analysis.
In the previous installment of this series, part 1, we set up a simple retrieval process that enabled us to get a single file from the JHU covid19 data archive on Github. (Before reading this tutorial, it might be helpful to read part 1, since this tutorial will draw heavily from it.)
So in part 1, we retrieved the data for covid19 confirmed cases and wrangled it into shape.
That was a good start, but there are actually two additional files that we’ll want to use as well … datasets for “deaths” and “recovered” cases.
We’ll want to get all three of those files, and we’ll ultimately need to combine them together into a single file. This will make our analyses much easier.
The process of getting the other two files will be fairly similar to getting the file for the “confirmed” cases. Because there are so many similarities, we’re going to create a group of functions that will enable us to operate on each of the three files.
Essentially, we’re going to set up a repeatable process that we can use and re-use in order to retrieve and wrangle our covid19 data.
Our general process
To do all of this, we’re going to build a set of individual functions that will do individual pieces of the wrangling process.
We’ll later combine those functions into a “master” function that we’ll be able to execute. This finalized function will retrieve and wrangle our data.
So essentially, we’re going to do this in pieces.
Here’s a rough overview of the process …
Table of Contents:
You’ll be able to use those links to skip to a particular point in the tutorial, but really, the tutorial will only make sense if you read the whole thing. I recommend that you read this from start to finish.
A quick note
One other quick note before we get started.
The functions and code here are intended to be “good enough,” but not necessarily perfect.
I’m writing these functions with two things in mind: baseline effectiveness and clarity.
First, I want the functions to be good enough to “get the job done” so we can get our data and manipulate it into shape.
Second, I want these functions to be relatively easy to understand for R students at a high-beginner to intermediate level.
That being said, we’re not going to be doing things like checking for valid function inputs, error handling, etc. I know that those things are important, but again, I just want these to work, but I still want them to be easy to read and understand for relative beginners.
… we’re just doing a simple tutorial here, not writing production code for MegaCorp, Inc.
R functions to get and wrangle covid19 data
Ok, let’s actually get started with creating the code.
Import R packages
Before we actually write or execute any code, we’ll need to import a few important R packages.
We’ll be using R to do this, but in particular, we’ll be using the Tidyverse package and the lubridate package. Remember: Tidyverse actually contains several other packages as well, including dplyr (which we’ll be using heavily) and ggplot2.
#================ # IMPORT PACKAGES #================ library(tidyverse) library(lubridate)
Now that we have the packages imported, we can get to work.
Define function: rename columns
Next, we’ll define a function to rename the columns.
covid_rename_columns <- function(input_data){
input_data %>%
rename('subregion' = 'Province/State'
,'country' = 'Country/Region'
,'lat' = 'Lat'
,'long' = 'Long'
) ->
output_data
return(output_data)
}
Notice that this is taking an input dataset as an argument, input_data. Inside of the function, we’re using dplyr::rename() to rename some of the columns of the dataset.
The output of the rename() process is being saved as output_data, which we’re returning from the function. Essentially, the function will return the data with renamed variables.
Also, keep in mind: we’re not going to use these functions individually, one at a time.
We’re going to wrap them up into a “master function” that will execute all of them in sequence to process our data in one go.
That being said, we’ll move on to the next function.
Define function: pivot data
Here, we’re going to “pivot” our data into a new shape.
As we saw in part 1, the raw data has a separate column for each date.
Here’s a “glimpse” of the confirmed data:
Observations: 264 Variables: 102 $ `Province/State` NA, NA, NA, NA, NA, NA, NA, NA, "Australian Capital Territory", "New Sout… $ `Country/Region` "Afghanistan", "Albania", "Algeria", "Andorra", "Angola", "Antigua and Ba… $ Lat 33.0000, 41.1533, 28.0339, 42.5063, -11.2027, 17.0608, -38.4161, 40.0691,… $ Long 65.0000, 20.1683, 1.6596, 1.5218, 17.8739, -61.7964, -63.6167, 45.0382, 1… $ `1/22/20` 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0… $ `1/23/20` 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0… $ `1/24/20` 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0… $ `1/25/20` 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0… ... $ `4/27/20` 1703, 736, 3517, 743, 27, 24, 4003, 1808, 106, 3004, 28, 1033, 438, 214, … $ `4/28/20` 1828, 750, 3649, 743, 27, 24, 4127, 1867, 106, 3016, 28, 1034, 438, 218, …
Again, we have one column for every date.
That’s not what we want. We want the different dates to exist as values under one single “date” column.
To get the data into this appropriate shape, we need to transpose the data.
To do this, we will use the pivot_longer() function to pivot the data from wide shape to long shape.
covid_pivot_data <- function(input_data, value_var_name){
input_data %>%
pivot_longer(cols = -one_of('country','subregion','lat','long')
,names_to = 'date'
,values_to = value_var_name
) ->
output_data
return(output_data)
}
Notice that we’re setting this function up with an input parameter called input_data. This will be the input dataframe.
We also have another input parameter called value_var_name. This will be the name of the new “values” column.
Inside the function, we’re using pivot_longer() to pivot all of the separate “date” variables into a single variable called date. We’re also creating a “values” variable, the name of which is provided by value_var_name. You’ll see later, when we use the function, that we’ll be able to use this to create a new column called “confirmed” that will have confirmed cases.
Define function: convert dates
Next, we’ll define a function to convert the newly created date variable into a proper R date.
We saw this in part-1 of this tutorial series, when we initially imported the confirmed cases dataset. When we imported the data and wrangled it into shape, the dates in the date variable were initially character data, not proper R dates. So we needed to modify the character dates to turn them into R dates.
We’ll do the same thing here.
Here, we’re going to define a function that will transform the “dates” in the date variable from character-based dates into proper R dates.
#======================
# DEFINE: Convert Dates
#======================
covid_convert_dates <- function(input_data){
input_data %>%
mutate(date = mdy(date)) ->
output_data
return(output_data)
}
The function accepts a dataframe as an input that we’re calling input_data.
Inside the function, we’re using the dplyr::mutate() to modify our date variable. Remember that the mutate() function creates new variables in a dataframe, but we can also use it to modify an existing variable. That’s what we’re doing here.
Specifically, inside of mutate, we’re using the mdy() function from lubridate, which transforms character based data in the form of MONTH-DAY-YEAR into an R date. Here, mdy() will take the date variable as an input, and store the output back into the date variable. Essentially, we’re overwriting the character-based dates in the date variable with R dates.
The output of mutate is being stored with the name output_data, and the function returns output_data.
So ultimately, the function will accept a dataframe, and output a new dataframe, and the date variable will be transformed from character to R date.
Define function: rearrange data
Next, we’ll define a function to rearrange the variables.
#=======================
# DEFINE: Rearrange Data
#=======================
covid_rearrange_data <- function(input_data){
input_data %>%
select(country, subregion, date, lat, long, everything()) %>%
arrange(country, subregion, date) ->
output_data
return(output_data)
}
Here, once again, the function has an input parameter called input_data that accepts a dataframe.
Inside the function, we’re using the dplyr::select() function to rearrange the columns. At a high level, the select() function enables you to “select” columns from a dataframe, but in this case, we’re actually using it to reorder the columns. Essentially, we are “selecting” the columns, but we’re selecting them in a new order.
After the select() function, we’re using the dplyr::arrange() to sort the rows of the dataframe. Here, we’re sorting by country, subregion, and date.
The output of this chain of operations is being sent to output_data, which is then returned by the function.
So the covid_rearrange_data() function takes a dataframe as an input, reorders the columns, sorts the rows, and then returns the re-arranged dataframe.
Define function: get and wrangle data
Okay …. we’re almost done.
Now, we’re going to take all of the functions that we’ve created so far, and wrap them up into a master data-wrangling function that will get and wrangle our covid19 data.
Let me show you the code first, and then explain.
#======================================
# DEFINE DATA COVID PROCESSING FUNCTION
#======================================
covid_get_data <- function(input_url, value_var_name){
covid_data_inprocess <- read_csv(input_url)
covid_data_inprocess <- covid_rename_columns(covid_data_inprocess)
covid_data_inprocess <- covid_pivot_data(covid_data_inprocess, value_var_name)
covid_data_inprocess <- covid_convert_dates(covid_data_inprocess)
covid_data_inprocess <- covid_rearrange_data(covid_data_inprocess)
return(covid_data_inprocess)
}
So what are we doing here?
This new function, covid_get_data(), just combines the previous functions that we defined. It accepts an input URL as the first argument, and we can provide a "value_var_name" name as the second argument.
When we use this in a moment, you'll see that this will enable us to process the different covid19 files – the files for confirmed, dead, and recovered cases – differently.
Get data
Let's actually use our new function, so you can see how it's used.
Here, we'll first define URL locations for different data files: url_confirmed, url_deaths, and url_recovered. These are the locations of the covid19 csv files that are compiled by JHU.
url_confirmed = 'https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_confirmed_global.csv' url_deaths = 'https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_deaths_global.csv' url_recovered = 'https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_recovered_global.csv'
Next, we'll use the covid_get_data() to retrieve the different files at those locations:
covid_confirmed = covid_get_data(url_confirmed,'confirmed') covid_deaths = covid_get_data(url_deaths,'dead') covid_recovered = covid_get_data(url_recovered,'recovered')
After running the process, we have 3 different dataframes: covid_confirmed, covid_deaths, and covid_recovered.
Print records
Quickly, let's print out some records.
Here, we'll print some records from the new covid_deaths dataframe.
print(covid_deaths)
OUT:
# A tibble: 26,866 x 6 country subregion date lat long dead1 Afghanistan NA 2020-01-22 33 65 0 2 Afghanistan NA 2020-01-23 33 65 0 3 Afghanistan NA 2020-01-24 33 65 0 4 Afghanistan NA 2020-01-25 33 65 0 5 Afghanistan NA 2020-01-26 33 65 0 6 Afghanistan NA 2020-01-27 33 65 0 7 Afghanistan NA 2020-01-28 33 65 0 8 Afghanistan NA 2020-01-29 33 65 0 9 Afghanistan NA 2020-01-30 33 65 0 10 Afghanistan NA 2020-01-31 33 65 0 # … with 26,856 more rows
Notice that the data has country, subregion, date, etc ... but it also has a column for "dead".
We set up covid_get_data() to create this variable when we called the function with the code covid_get_data(url_deaths,'dead').
I'll let you print the other files ... you'll see that they're structured in a similar way, but with columns for confirmed and recovered respectively.
Get record counts
Quickly, let's get the record counts for each file.
We want these, because the next step is merging together the files, and we can use the record counts before and after as a "check" to make sure that the join worked properly.
count(covid_confirmed) count(covid_deaths) count(covid_recovered)
Currently, as of May 2, the covid_confirmed dataset has 26866 records and so does the covid_deaths data. The covid_recovered file has slightly less with 25452.
Merge data
Now, we're going to merge the three files together.
Before we do this, we'll drop some columns that would be duplicates.
Drop unnecessary columns
Here, we're going to delete unnecessary columns.
#------------------------- # DROP UNNECESSARY COLUMNS #------------------------- covid_deaths <- covid_deaths %>% select(-long, -lat) covid_recovered <- covid_recovered %>% select(-long, -lat)
To do this, we're just using the select() method to remove individual columns.
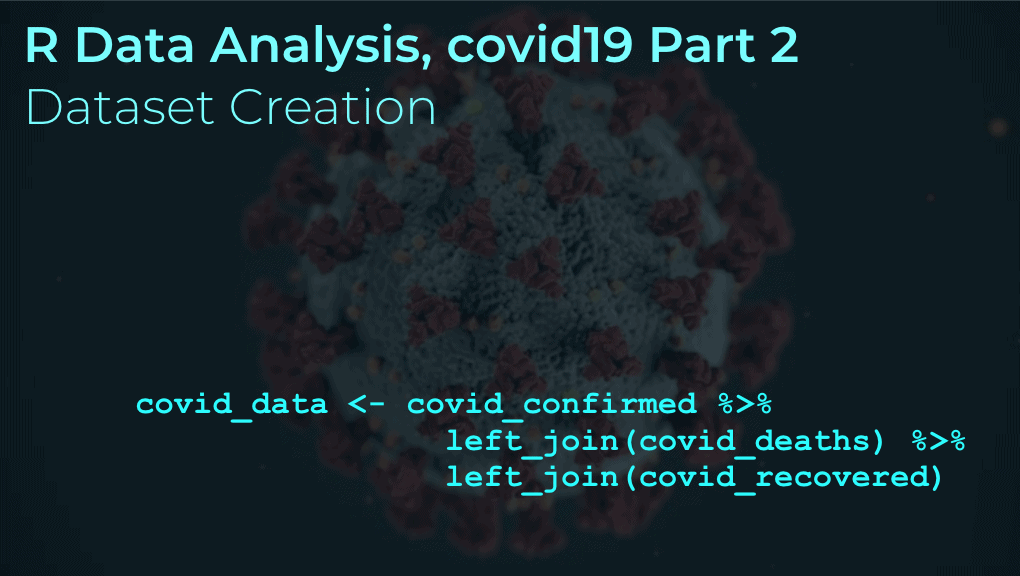
Merge data
Now that we have our three files, let's merge them together.
Here, we'll use left_join() to join the three files together on country, subregion, and date.
covid_confirmed %>% left_join(covid_deaths, on = c(country, subregion, date)) %>% left_join(covid_recovered, on = c(country, subregion, date)) -> covid_data
We're saving the output as covid_data, which is a dataframe that has covid19 confirmed cases, deaths, and recovered cases, by country, subregion, and date.
Print records
Let's just print some of the data, so we can see it:
print(covid_data)
OUT:
# A tibble: 26,866 x 8 country subregion date lat long confirmed dead recovered1 Afghanistan NA 2020-01-22 33 65 0 0 0 2 Afghanistan NA 2020-01-23 33 65 0 0 0 3 Afghanistan NA 2020-01-24 33 65 0 0 0 4 Afghanistan NA 2020-01-25 33 65 0 0 0 5 Afghanistan NA 2020-01-26 33 65 0 0 0 6 Afghanistan NA 2020-01-27 33 65 0 0 0 7 Afghanistan NA 2020-01-28 33 65 0 0 0 8 Afghanistan NA 2020-01-29 33 65 0 0 0 9 Afghanistan NA 2020-01-30 33 65 0 0 0 10 Afghanistan NA 2020-01-31 33 65 0 0 0 # … with 26,856 more rows
As you can see, covid_data has covid19 confirmed cases, deaths, and recovered cases, by country, subregion, and date. It also has the latitude and longitude of the country/subregion.
Add new cases
Finally, we're going to add the daily new confirmed cases as a new variable.
This variable will be called new_cases, and it will equal to the daily change in confirmed cases.
In particular, we need to do this within a country/subregion. For example, we want to calculate daily new cases for the US separately from the daily cases for other countries.
This is not terribly complicated, but we need to be careful.
To do this, we need to group our data by country and subregion.
The data also need to be sorted by country, subregion, and date. We're doing this with dplyr::arrange().
And ultimately, we're using dplyr::mutate() to add the new variable called new_cases, which is equal to the confirmed cases, minus the confirmed cases on the previous day (but within a given country/subregion!).
covid_data %>% arrange(country, subregion, date) %>% group_by(country, subregion) %>% mutate(new_cases = confirmed - lag(confirmed)) %>% ungroup() -> covid_data
We're saving the output back to covid_data, which means that we're overwriting the dataset.
Be careful!
Whenever you have a risk of overwriting your data, check your code. Having said that, I checked this, and it should be okay.
Examine final data
Let's take one last look at our finalized dataset.
print(covid_data)
OUT:
# A tibble: 26,866 x 9 country subregion date lat long confirmed dead recovered new_cases1 Afghanistan NA 2020-01-22 33 65 0 0 0 NA 2 Afghanistan NA 2020-01-23 33 65 0 0 0 0 3 Afghanistan NA 2020-01-24 33 65 0 0 0 0 4 Afghanistan NA 2020-01-25 33 65 0 0 0 0 5 Afghanistan NA 2020-01-26 33 65 0 0 0 0 6 Afghanistan NA 2020-01-27 33 65 0 0 0 0 7 Afghanistan NA 2020-01-28 33 65 0 0 0 0 8 Afghanistan NA 2020-01-29 33 65 0 0 0 0 9 Afghanistan NA 2020-01-30 33 65 0 0 0 0 10 Afghanistan NA 2020-01-31 33 65 0 0 0 0 # … with 26,856 more rows
As you can see, our covid_data dataset has confirmed cases, deaths, recovered cases, and daily new cases by country, subregion, and date.
This is a file that we can analyze and visualize.
One quick note: if you run this, your data may be different
One final note.
If you run this code yourself, any time after May 3, your data will have more rows.
That's because Johns Hopkins is updating their database roughly every day (at least as of the time of posting).
So if you run this code after May 3, you'll have more data ... data for the days after May 3.
Other than that though, your data should be very similar.
Next steps
So we finally have a working covid19 data file.
Here, we've done most of the hard work of retrieving and wrangling our data. We created a repeatable process (using R functions and dplyr). And we've used that process to create a somewhat finalized covid19 dataset.
Having said that, there's still more to do.
I want to explore this data to check it. Here in this specific blog post, we created a process to create the data, but we should still check the data against some official numbers.
To do that, we'll need to take some subsets and compare against official data. We'll probably do that in the next tutorial.
After that, there are a lot of data visualizations that we can create with this data.
So in a few subsequent tutorials, we'll create some specific data visualizations using ggplot2.
Is there anything else you want to see in this R tutorial series?
Leave a comment in the comments section below and tell me.
Sign up to learn more
Do you want to see part 3 and the other tutorials in this series?
Sign up for our email list now.
When you sign up, you'll get our tutorials delivered directly to your inbox.
Thank you for offering us your expertise and tutoring.
You’re welcome
Great insights! I follow these tutorials in R thoroughly. Thanks
Good to hear …. thanks for reading JB
Thank you so much for sharing such an insightful coding techniques of organizing datasets into a coherent one. I really enjoy working with R than Python, because I started data analysis with R, I am addicted to R. The skills I acquired from these tutorials will greatly help my career. Thank you so much for generously sharing with us.
You’re welcome.
Great to hear that these tutorials will help you in your career …
my suggestion is to perform some exploratory statistics analysis…compare the values between countries
This is only part 2.
We’ll get to all of that.
I am getting the following numbers which do not match with yours.
Kindly advise where I must have gone wrong
head(covid_data)
# A tibble: 6 x 8
country subregion date lat long confirmed dead recovered
1 Afghanistan NA 2020-01-22 33 65 0 0 0
2 Afghanistan NA 2020-01-23 33 65 0 0 0
3 Afghanistan NA 2020-01-24 33 65 0 0 0
4 Afghanistan NA 2020-01-25 33 65 0 0 0
5 Afghanistan NA 2020-01-26 33 65 0 0 0
6 Afghanistan NA 2020-01-27 33 65 0 0 0
> dim(covid_data)
[1] 28728 8
We’re retrieving the data from a datastore on Github that’s managed by Johns Hopkins.
The Github data store is being updated once per day with new data.
I ran my code around May 3.
If you ran your code on or around May 9 (when you posted your comment), then you’ll have more data than I showed in the tutorial.
I did not mention that when I initially posted the tutorial, but I’ve updated the tutorial with a section that explains this.
Thanks a lot. Relieved that I am not wrong
Hi, what is the “ungroup()” for?
It’s to undo the
group_by()that grouped the data when we calculated the new cases.We needed that grouping when we computed new cases.
But having that grouping could mess up other calculations in the future, so we needed to remove it after we were finished using it.